
 By Nathanael Caveney, PhD Candidate, Strynadka Lab & Jeffrey To, Graduate Student, Kim Lab
By Nathanael Caveney, PhD Candidate, Strynadka Lab & Jeffrey To, Graduate Student, Kim Lab
Antibiotic resistance has become a major issue in recent years, and is believed to be directly linked to the livestock industry which accounts for ~80% of antibiotic use in North America. Frighteningly, this is expected to rise by nearly 70% globally by 2030 (Van Boeckel et al., PNAS, 2015). Coupled to its widespread and rising use, the development of novel antibiotics is in great decline, with no new classes of antibiotic having entered the clinic in the last 30 years (Silver, Clin Microbiol Rev, 2011). Since the majority of antibiotics in the clinic only inhibit a small subset of the potential bacterial targets, a major focus in the field is to discover and characterize novel targets.
The later stages of bacterial cell wall synthesis, have traditionally been targeted for antibacterial development, with over 50% of the antibiotics prescribed in the clinic designed to disrupt bacterial cell wall crosslinking. These approaches have been successful due to the importance of the cell wall in the strength and viability of bacteria, as well as having no counterpart in mammals. Therefore, it is of interest to pursue novel targets of bacterial cell wall biosynthesis that occur at different points in this complex pathway. One such example is the targeting of lipid recycling pathways, which are critical in cell-wall biosynthesis.
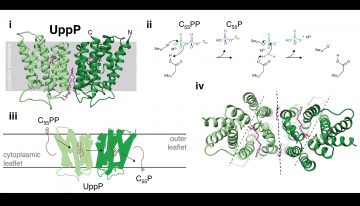
Lipid carrier synthesis and recycling. (i) Structure of UppP (green) in complex with structural lipid and substrate mimic, monoolein (pink) (E. coli, 6CB2). (ii) Proposed reaction mechanism for UppP mediated dephosphorylation of C55PP. (iii) Schematic representation of proposed UppP phosphatase-couple lipid flippase activity. (iv) Twofold symmetry axis of the UppP dimer (black) and twofold pseudosymmetry axes of each monomer (grey).
A recent study by Workman et al. (Nat Comm, 2018) has led to the structural characterization of a novel protein involved in this crucial bacterial process. The enzyme Undecaprenyl Pyrophosphate Phosphatase (UppP) is responsible for removing phosphate from both de novo synthesized carrier lipids and recycled lipids that have already contributed to cell wall formation. Using in meso crystallography techniques, the structure of this integral membrane protein was solved to 2 Å resolution, allowing a deeper understanding of this potential antibiotic target.
In this work, the protein was found to have a novel topology consisting of inverted repeats and pseudosymmetry across a monomer of UppP. Detailed structural characterization provided additional insights into the mechanism of phosphatase function, which had previously been debated in the field for some time. In addition, this structure also has features reminiscent of many cross-membrane transporters and therefore proposes the intriguing possibility that UppP may have flippase activity coupled to its phosphatase activity.
This structure opens the door for the development of novel structure-guided antibiotics. Successful design and introduction of UppP inhibitors will assist in the fight against the current resistance crisis and lead to further understanding of these lesser studied aspects of cell wall biosynthesis. Further research is needed to explore the potential role of UppP as a flippase in order to provide a conclusive verdict on UppP’s role in these processes, as well as a greater insight into bacterial cell wall development.
- Van Boeckel, T. P. et al. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5649–54 (2015).
- Silver, L. L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24, 71–109 (2011).
- Workman, S. D., Worrall, L. J. & Strynadka, N. C. J. Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling. Nat. Commun. 9, 1159 (2018).
- Caveney, N. A., Li, F. K. & Strynadka, N. C. Enzyme structures of the bacterial peptidoglycan and wall teichoic acid biogenesis pathways. Curr. Opin. Struct. Biol. 53, 45–58 (2018).


