 Written by: Erik Lamoureux, PhD Student, Ma Lab (left)
Written by: Erik Lamoureux, PhD Student, Ma Lab (left)
Edited by: Alexandra Witt, MSc Student, Pryzdial Lab (right)
This post was written by Erik Lamoureux, an PhD Student from the Ma Lab (Multi-Scale Design Laboratory) at the Centre for Blood Research (CBR) at the University of British Columbia.
This is a summary of his lab’s paper “Assessing red blood cell deformability from microscopy images using deep learning”, published in the journal Lab on a Chip.
Read a version of this story on the Canadian Blood Services’s Research. Education. Discovery. (R.E.D.) blog: Emerging Artificial Intelligence Methods to Assess Transfusion Quality.
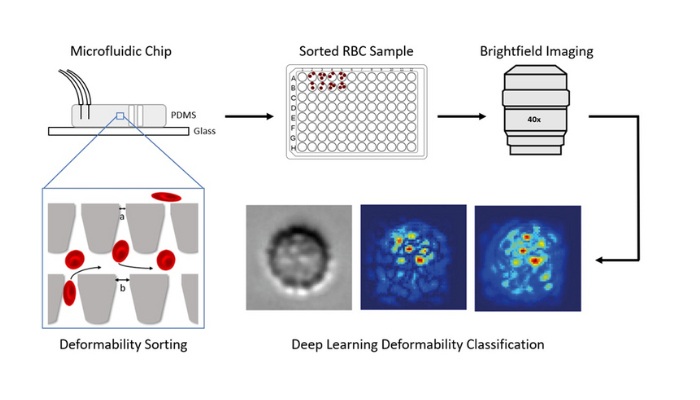
Fig. 1. Experimental procedure. Red blood cells are sorted based on their deformability using a microfluidic device. The sorted cells are extracted, imaged, processed, and assessed using artificial intelligence (AI) image analysis methods. This AI method, called deep learning, can accurately predict a donor’s deformability profile from cell images 1.
Around half of all Canadians will require a blood transfusion in their lifetime or know someone else who will. Despite how common blood transfusions are, only four percent of Canadians are blood donors2. Due to the scarcity of blood for transfusion, it is vital to use the existing blood supply as efficiently as possible. It is well known that blood type is assessed – O, A, B, or AB, with +/- Rhesus (Rh) factor – to ensure blood compatibility between the donor and recipient. In addition to blood type, recent research indicates that the shape and deformability of red blood cells (RBCs) are important factors contributing to the effectiveness of a blood transfusion.
Red blood cells and deformability
The deformability of red blood cells – their ability to squeeze through small spaces – is vital for their navigation through the body. During circulation, RBCs squeeze through small gaps in the microvasculature to deliver oxygen and remove carbon dioxide from cells and tissues. Not only do RBCs need to squeeze through these small gaps, but they must also do this repeatedly; they circulate through the body once every 60 seconds and endure for a 120-day life span. Due to this, RBC deformability is vital for effective circulation.
Why deformability matters
Red blood cell deformability degrades as the cell ages, and when the cell becomes too rigid to pass through the microvasculature, it is engulfed and destroyed by immune cells called phagocytes. Because of this relationship between a cell’s deformability and longevity, RBC deformability is considered a potential biomarker for transfusion quality. Donated RBCs for transfusion confer differing benefits to transfusion recipients3, 4, potentially in part due to natural differences in RBC deformability between donors 5. Ideally, blood banks could sort blood units based on patient need. More deformable and longer-lasting blood should be distributed to chronic transfusion patients to extend the time between their needed transfusions. On the other hand, more rigid and shorter-lasting blood should be distributed to acute transfusion patients (e.g., trauma patients) who just need a blood top-up and can otherwise produce an adequate blood supply.
Microfluidics to assess deformability
In our lab, we developed a microfluidic device to sort RBCs based on their deformability6. RBCs suspended in fluid are sent through microchannels that are as small as the width of a human hair. Microfluidic platforms provide advantages compared to conventional laboratory assessment techniques. Namely, microfluidics benefit from precise fluid manipulation and control, require small volumes of fluid and biological samples (on the order of fractions of a millilitre), and can probe features of individual cells in a fast and highly efficient manner. In our device, the cells enter a grid-like sorting region consisting of small constrictions of different sizes for the cells to squeeze through. This sorting is similar to cells squeezing through small gaps in the body’s microvasculature. The cells are sorted from the bottom of the grid to the top (relative to Fig. 1) and these constrictions get progressively smaller along the flow path. Based on a cell’s ability to squeeze through these gaps, they are sorted to different outlets indicating their level of deformability. This allows us to determine a donor-specific rigidity score and provides us with the true deformability levels of the cells for use in training a machine learning model.
Artificial intelligence (AI) to assess deformability
To increase the accessibility of determining RBC deformability, we sought to develop computer-based techniques to predict deformability from images. After microfluidic deformability sorting, sorted cells are extracted from the device and imaged using a microscope. The microscope captures digital images of the cells which are then processed into smaller single-cell images. These single cell images are input to an artificial intelligence model called a deep learning network. This network consists of cascading and interconnected mathematical operations that learn patterns in the data. The network updates its connections and how the data flows through these structures based on feedback from the microfluidic deformability labels corresponding to each image. By passing enough images through the network many times, the model is trained to identify a cell’s deformability level based on its surface features, patterns, and structures. All told, this AI-based method correctly predicted the deformability of individual RBC images with 81 ± 11% accuracy averaged across 10 donors1. As both methods also produce a rigidity score for the RBCs of each donor, we further assessed the model by comparing the AI-derived rigidity score to the ex vivo-derived score. We found that the deep learning-derived rigidity scores were accurate to within 10 ± 7% of the value obtained using the microfluidic device.
Benefits of artificial intelligence methods
Using machine learning methods to determine red blood cell properties from images has huge potential to benefit transfusion medicine research and practice7. By assessing red cell quality based on morphological features identified in images, this approach could be introduced to any research laboratory or clinic with an adequate microscope and computer. Future work will focus on streamlining this procedure, determining viable use cases and limitations, and ideally developing a generalizable model that can assess the RBC deformability profile of new donors at the point of care.
References
1 E. S. Lamoureux, E. Islamzada, M. V. J. Wiens, K. Matthews, S. P. Duffy, and H. Ma, ‘Assessing red blood cell deformability from microscopy images using deep learning’, Lab Chip, vol. 22, no. 1, pp. 26–39, 2022, doi: 10.1039/D1LC01006A.
2 Canadian Blood Services, ‘Half of all Canadians will either need blood or know someone who will need blood at some point in their lives.’, Canadian Blood Services, 2022. [Online]. Available: https://myaccount.blood.ca/en/fact/half-all-canadians-will-either-need-blood-or-know-someone-who-will-need-blood-some-point-their
3 O. Garraud and J.-D. Tissot, ‘Blood and Blood Components: From Similarities to Differences’, Front. Med., vol. 5, p. 84, Apr. 2018, doi: 10.3389/fmed.2018.00084.
4 J. R. Hess, ‘Conventional blood banking and blood component storage regulation: opportunities for improvement’, Blood Transfusion, 2010, doi: 10.2450/2010.003S.
5 E. Islamzada et al., ‘Deformability based sorting of stored red blood cells reveals donor-dependent aging curves’, Lab Chip, vol. 20, no. 2, pp. 226–235, 2020, doi: 10.1039/C9LC01058K.
6 Q. Guo et al., ‘Deformability based sorting of red blood cells improves diagnostic sensitivity for malaria caused by Plasmodium falciparum’, Lab Chip, vol. 16, no. 4, pp. 645–654, 2016, doi: 10.1039/C5LC01248A.
7 J. A. Sebastian, M. C. Kolios, and J. P. Acker, ‘Emerging use of machine learning and advanced technologies to assess red cell quality’, Transfusion and Apheresis Science, vol. 59, no. 6, p. 103020, Dec. 2020, doi: 10.1016/j.transci.2020.103020.


