By Prashant Kumar, PhD Candidate at Kizhakkedathu Lab, CBR
By Prashant Kumar, PhD Candidate at Kizhakkedathu Lab, CBR
With an advent of bacterial resistance, it has become increasingly important to understand the mechanism involved in the multi-drug resistance of so-called “superbugs”. Staphylococcus aureus (S. aureus) is a common bacteria found on the skin of healthy individuals. S. aureus is normally harmless; however, it may cause minor skin infections such as pimples/boils upon penetration of the epidermus. It can also lead to serious infections in the bloodstream. Methicillin antibiotic is a common treatment for S. aureus infections. However, some strains of S. aureus, such as Methicillin Resistant S. aureus (MRSA) are not susceptible to methicillin leading to devastating consequences in the clinic.
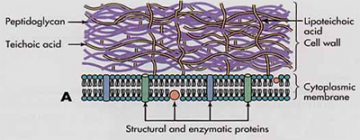
S. aureus is a Gram positive bacterium and possesses a thick layer of teichoic acids (TAs) composed of carbohydrate chains. TAs attached to the bacterial cell wall, called WTAs, eventually form cross-stitched matrices that protect Gram positive bacteria and allow for growth and survival in severe environments (see image). Although the exact role of TAs is not fully understood, these polymers compose up to 60% of the cell wall, with bacteria investing vast amounts of energy to their construction and maintenance. Previous studies have demonstrated that TAs play a role in antimicrobial resistance, induction of inflammation and biofilm formation. Many of these processes depend on the attachment of sugar groups to TA (Figure 2), mediated by dedicated enzyme called glycosyltransferase. Recently, it has been also shown that attachment of sugar groups to WTA is also involved in MRSA resistance.
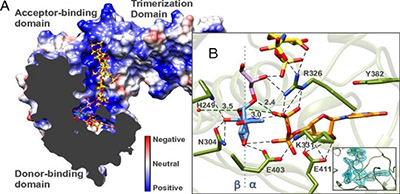
A recent PNAS publication by Dr. Solmaz Sobhanifar and Dr. Liam James Worrall from the Strynadka laboratory at CBR present the structure and mechanism of the S. aureus WTA glycosyltransferase enzyme – called TarM. The authors solved the X-ray crystal structure by forming a complex between TarM and its substrates. They used catalytic mutants of TarM and modeling of the ternary complex to elucidate an internal nucleophilic substitution-like reaction mechanism.
The interplay between TarM and a related glycosyltransferase enzyme, TarS, are responsible for the pattern of the sugar attachment to teichoic acids. Considering the configuration and extent of the glycosidic linkage varies with certain strains of S. aureus, this glycosylation pattern seems to be important in host immune response, phage binding and methicillin resistance.
It is clear that the ‘glycocode’ (complex glycosylation process) plays a vital role in bacterial survival and pathogenicity. The authors believe that TarM and TarS would be ‘lucrative targets for novel therapeutic agents in light of an ever-decreasing antibiotic arsenal’.
This research was funded by the CIHR (Banting), the MSFHR Fellowship program, the Howard Hughes Medical Institute International Scholar program and the BC Knowledge Development Fund.