Low levels of drug resistant HIV can emerge when exposed to drugs. Researchers at the BC Centre for Excellence in HIV/AIDS have developed a “deep” sequencing approach that gives an in-depth cross-section of the HIV envelope profile in patients.
HIV is extracted from patient blood samples and a portion of the envelope gene (which mediates HIV entry) is amplified. This region, called V3, encodes mutations which allow a virus to enter cells using one of two proteins, called CCR5 or CXCR4. Maraviroc disrupts the interaction of HIV with CCR5, but has poor activity against viruses that use CXCR4. Thus, a tropism assay is needed to determine which protein a patient’s HIV uses to enter cells.
Snapshot of a deep sequencing run in progress. When nucleotide bases incorporate into the DNA strand, they are identified by a flash of light.
The deep sequencing tropism assay is performed by pyrosequencing on a 454 Genome Sequencer FLX (“454”), shown below. Each sample generates thousands of sequences, representing the viral population that exists in a patient’s blood. The sequences can then be analyzed with bioinformatic algorithms to determine what proportion of them is resistant to maraviroc.
As recently reported in the journal Clinical Infectious Diseases (2011;53(7):732–742), Luke Swenson and colleagues in the lab of CBR member Dr. Richard Harrigan, tested this approach in a large clinical trial of maraviroc in patients beginning antiretroviral therapy, called the MERIT trial. Patients re-screened as having insignificant levels of resistant virus according to deep sequencing had better treatment responses than those with resistance. A higher proportion of the patients without resistance had undetectable HIV viral loads in their blood on treatment, shown below. The 454 deep sequencing approach also had similar performance to another commonly used screening assay called ESTA.
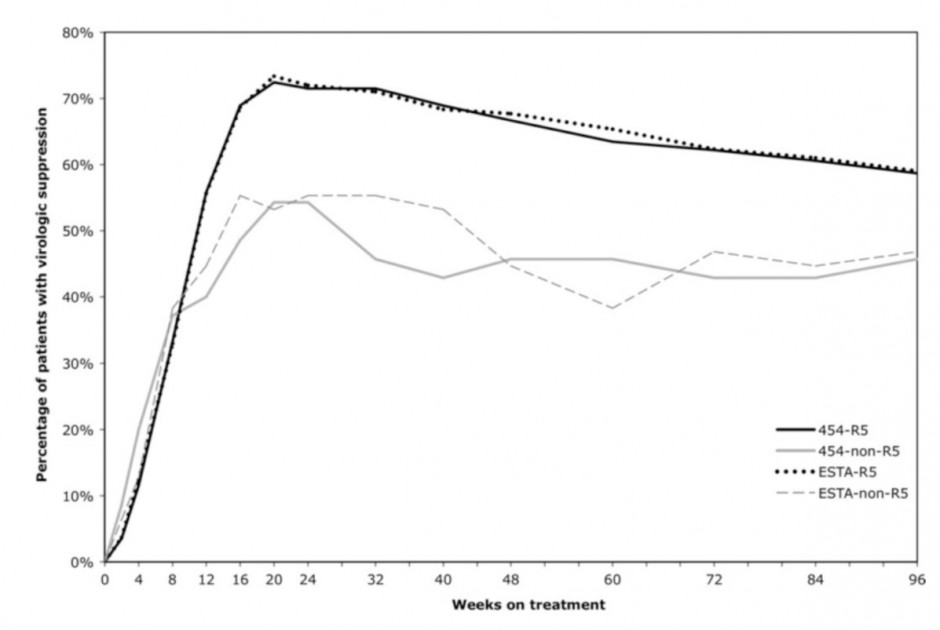
Deep sequencing of the V3 region of the HIV envelope gene gives a high-resolution cross section of the viral population in a patient’s blood. By looking for low levels of virus resistant to maraviroc, this approach can better predict suppression of HIV by combination drug therapy.
Read the whole story by clicking here.



Comments are closed, but trackbacks and pingbacks are open.