 By Sean Workman, MSc Student, Strynadka Lab, CBR
By Sean Workman, MSc Student, Strynadka Lab, CBR
Sialic acid sugars are a group of carbohydrates that are found in a wide variety of organisms, from simple bacteria to complex eukaryotes such as humans. Polysialic acid (polySia) is an extended polymer of sialic acid that is transferred to very few but specific protein substrates such as neural cell adhesion molecule (NCAM) in the vertebrate neural system. Modification of NCAM with polySia promotes migration of cells, a process that is critical for both the development of the embryonic brain and neural plasticity of the adult brain. Interestingly, elevated levels of polySia have been implicated in the malignant potential of tumors, tumor metastasis and poor clinical prognosis.
In eukaryotes, polysialylation is catalyzed by members of the St8Sia subfamily of sialyltransferases (STs). In this process, a polyST transfers a sialic acid from a donor molecule to the terminal sialic acid on an acceptor molecule one sialic acid at a time to generate the polySia polymer. While several monoSTs from this family have been characterized in terms of their cellular distributions and acceptor preference, the molecular details of how polySTs function and interact with acceptor proteins such as NCAM have remained unclear. In a recent paper published in Nature Structural and Molecular Biology, postdoctoral fellow Gesa Volkers, together with members of the Strynadka Lab at the Centre for Blood Research, set out to uncover these details by solving the crystal structure of human St8SiaIII, a close homolog of the most clinically important human polyST, ST8SiaIV.
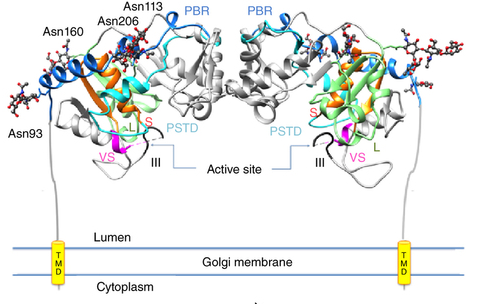
Overall structure of human ST8SiaIII with various motifs highlighted. Source: Volkers et al. Nature Structural & Molecular Biology 22 (2015).
Because the identity of the ST8SiaIII acceptor molecule was unknown, the enzyme was assayed on the mammalian glycan array at the Centre for Functional Glycomics, using a sophisticated protocol for STs. This analysis revealed that ST8SiaIII has a preference for a novel sulphated sialylated glycan acceptor. Members of Dr. Stephen Withers’ lab succeeded in synthesizing the acceptor, which was then used to obtain detailed kinetic information on the enzyme. The synthesized acceptor was also used to obtain the crystal structure of the “ternary complex” with the enzyme bound to both donor and acceptor molecules. This was the first structure of a polyST solved from any species, shedding light on the molecular basis for acceptor binding and leading to the identification of novel domains important for polyST activity. Structural analysis of St8SiaIII also helped identify individual amino acids important in the proposed enzymatic mechanism, and rationalized the difference in acceptor preference between this polyST and previously characterized monoSTs.
The work in this paper has significantly increased our understanding of how these important enzymes work and opens the door for further research into the development of anti-cancer drugs targeting tumor metastasis.



