 By Lily Takeuchi, Graduate Student, Kizhakkedathu Lab
By Lily Takeuchi, Graduate Student, Kizhakkedathu Lab
Part 1 of 2 – Please note, this post is intended for an audience with a strong scientific background.
In September 2017, a study conducted by researchers at Sun Yat-sen University (Guangzhou, China) demonstrated the feasibility of curing genetic diseases in human embryos through the use of a base editor system to correct β-thalassemia mutant genes. Though still far from reaching repair rates adequate for clinical implementation, the research has garnered significant international attention and concern. As research in germline editing reaches new and exciting frontiers, society is prompted to decide how science can act synergistically with policy to evaluate ethical implications and advance our knowledge of genetic engineering. The following article is part one of a two part series on the science and ethics of germline engineering.
The manipulation of “defective” genes in the human embryo could predictably result in a cure that could be retained through generations to come. Yet, this approach also yields the potential to immortalize unforeseen consequences in an indestructible vehicle: a human genome. Published in Protein & Cell, Liang et al. aimed to demonstrate the feasibility of using base editing systems in pursuit of curing genetic disease in human embryos. The base editing system, uses a RNA-protein complex developed at Harvard (see Fig 1). Unlike the guide-and-cut approach of CRISPR-Cas9 editing, base editing harnesses chemical conversion of bases without introducing double stranded breaks to correct mutations. The protein component deaminates cytidine to uridine (C → U) resulting in either a C → T or G → A base conversion in the target gene.
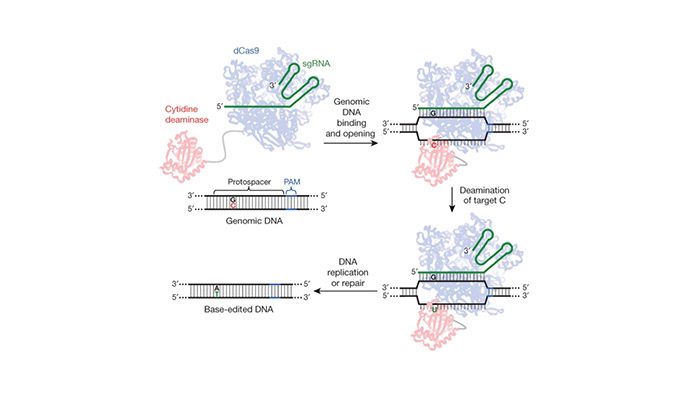
Figure 1: Gene editing without double stranded breaks. Base editing system using a Cas9-cytidine deaminase fusion protein mediates guide-RNA directed C → U conversion in the target genomic sequence (from AC Komor et al. Nature. 2016, 533, 420 doi:10.1038/nature17946)
Liang et al. utilized this base editing system to correct a mutation in the gene coding for β-hemoglobin (HBB). β-thalassemias are a group of inherited blood disorders of abnormal synthesis of the β-hemoglobin chain. β-thalassemia can be caused by single base substitutions upstream of the β-hemoglobin gene, a common defect being a −28 A → G mutation in HBB. The phenotypic presentation is an underproduction of adult hemoglobin resulting in a need for regular blood transfusions to sustain life.
Testing the system in vitro with patient-derived skin fibroblasts demonstrated a repair success of up to 17.8%. The researchers next advanced to testing these methods using lab-grown human embryos. Development of the embryos was done using a nuclear transfer method: after removal of the nuclear material from human oocytes, the embryos were fused with nuclei harvested from lymphocytes of β-thalassemia patients. The base editing system and a guide RNA targeting sequence was then injected into the cytoplasm. The group reported a repair efficiency up to 25.9% in resultant blastomeres 48-hours after introduction of the base editing machinery.
Although these embryos were non-viable and the technology remains far from achieving repair rates adequate enough for clinical implementation, the controversial research has introduced new dilemmas to be considered. In early stages of study, base editing has been demonstrated to be unpredictable and imperfect. The very technology that wields the power to cure can also create pathology, the inheritable permanence of which raises numerous questions: what are the consequences of introducing new mutations into the gene pool? What happens to embryos that go uncorrected? Given the stakes, how, and to what extent should caution be practiced? Click here to read Part 2 of this series!
References
- Liang, P.; Ding, C.; Sun, H.; Xie, X.; Xu, Y.; Zhang, X.; Sun, Y.; Xiong, Y.; Ma, W.; Liu, Y.; Wang, Y.; Fang, J.; Liu, D.; Songyang, Z.; Zhou, C.; Huang, J. Correction of β-thalassemia mutant by base editor in human embryos. Protein and Cell. 2017, 8, 11, 811-822.
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Xuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-strand DNA cleavage. Nature. 2016, 533, 420.
- Lanphier, E.; Urnov, F.; Haecker, S.R.; Werner, M. Smolenski, J. Don’t edit the human germ line. Nature Comment. 2015, 519, 410.


