 By Bronwyn Lyons, PhD Candidate, Strynadka Lab
By Bronwyn Lyons, PhD Candidate, Strynadka Lab
Certain bacteria, including the opportunistic pathogen Pseudomonas aeruginosa, use a mechanism of motility called swarming to travel along surfaces such as the mucosal lining of the lung1. This mode of transportation is made possible by long propeller-like tails called flagella, which allow for swift dispersion of the pathogen throughout the infected tissue. Swarming motility is also important for the establishment of bacterial biofilms. These matrix encapsulated communities of bacteria tend to be resistant to even aggressive antibiotic treatments. P. aeruginosa is a particularly problematic pathogen for immunocompromised patients, such as those with Cystic Fibrosis, where chronic and life-threatening infections often occur.
Members of the Hancock lab recently published a study detailing the multidrug adaptative antibiotic resistance of P. aeruginosa swarming cells2. Notably, Coleman and colleagues confirmed that swarming P. aeruginosa cells are capable of acquiring increased resistance to multiple antibiotics prior to previous exposure. They explored the genetic mechanisms behind this adaptive resistance, identifying key changes that occur in swarming cells at the transcriptome level.
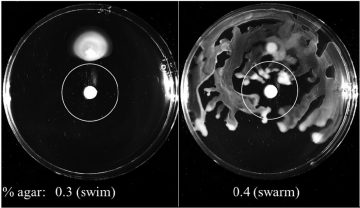
Figure 1: Illustration of Pseudomonas aeruginosa swimming (left) and swarming (right) motility on agar plates with tobramycin-soaked disks. Swarming bacteria approached closer to the centre, indicating heightened antibiotic resistance.
Coleman and colleagues explored the role of swarming in the development of resistance to a variety of antibiotics, including for example, ß-lactams (e.g. piperacillin) and aminoglycosides (e.g. tobramycin). Agar plates designed to mimic either swarming or swimming growth conditions were inoculated with bacterial culture. These experiments make use of the fact that swimming, another bacterial mechanism of motility that occurs in liquid environments rather than surfaces, is supported at lower percentages of agar than swarming. Antibiotic resistance was measured based on how close the swarming (or swimming) cells grew towards a filter paper disk soaked with the antibiotic of choice at the centre of the plate, with more resistant bacteria approaching closer to the centre Coleman and colleagues found that P. aeruginosa swarming cells not only exhibited heightened resistance to most antibiotic classes, as opposed to for example swimming cells (Figure 1), but also were more resistant to aminoglycoside antibiotics than they were towards ß-lactams.
So, what is the mechanism behind the heightened antibiotic resistance in swarming cells? Using a sophisticated technique called RNA sequencing (RNA-Seq), researchers in the Hancock Lab sought to identify changes in the regulation of specific genes in swarming cells when exposed to different antibiotics. Interestingly, swarming (versus swimming) cells exhibited differential expression of numerous RNA transcripts, corresponding to 28% of the entire P. aeruginosa genome. This suggests that antibiotic resistance under native swarming conditions is likely a cumulative effect of many genes. Furthermore, when treated with the antibiotic tobramycin, swarming cells exhibited significant upregulation in the expression of genes that correspond to proteins that are involved in drug efflux (designed to pump antibiotics out of the bacterial cell) and downregulation of those involved in membrane permeability. As this is only observed during antibiotic exposure, a different set of mechanisms of resistance must be at play during antibiotic exposure when swarming.
In summary, Coleman and colleagues have illustrated the dynamic and multi-dimensional role of bacterial motility and how it affects antibiotic resistance. During swarming motility, many genes are dysregulated, resulting in a state of antibiotic resistance. Furthering our understanding of antibiotic resistance, and the multiple factors that allow P. aeruginosa to cause serious infections in susceptible individuals, is extremely important for efficient and effective therapeutic design.
1 Kearns, D. (2010) A field guide to bacterial swarming motility. Nat. Rev. Micro. 8, 634-644.
2 Coleman, S.R., Blimkie, T., Falsafi, R., Hancock, R.E.W. (2020) Multidrug adaptive resistance of Pseudomonas aeruginosa swarming cells. Antimicrob. Agents Chemother. 64, e01999-19.


