 By Dr. Michael Hughes, Research Associate, McNagny Lab
By Dr. Michael Hughes, Research Associate, McNagny Lab
Thanks to a travel award from UBC’s Centre for Blood Research (CBR), I was thrilled to attend the Australian and New Zealand Society for Immunology 48th Annual Scientific Meeting (ASI 2019) in Adelaide, Australia. I was honoured to give a short talk at the meeting. This presentation was followed by plenty of opportunities to discuss my work with scientists interested in the gut microbiome-lung inflammation axis. Since most of these conversations were with trainees (i.e. the really smart people!) who rarely come to North America, these face-to-face connections would not have been possible without a travel award from CBR.

Photo: Adelaide Convention Centre along the “Karrawirra Parri” (which means redgum forest river)
Project background
There appears to be a very early window, perhaps only the first 3 months, in which the microbiome of human infants has a lifelong impact on their atopic susceptibility in later life1,2. In the McNagny lab, we can model the influence of early-life microbiota by treating pregnant mice with vancomycin in the drinking water (Fig 1). This antibiotic depletes gut bacteria responsible for producing short-chain fatty acids (SCFAs). The selective gut dysbiosis is presumably passed from mother to offspring as they nurse and grow, resulting in adult offspring mice with an exaggerated response to lung allergens like the house dust mite3. Intriguingly, we can “fix” the exaggerated allergic response by providing the missing SCFAs in the drinking water.
At ASI 2019, I presented data suggesting that metabolites from SCFA-producing gut bacteria alter repopulating hematopoietic stem and progenitor cells (HSPCs), resulting in long-term immune skewing. I highlighted bone marrow (BM) transplant experiments showing how the atopic phenotype from dysbiotic mice can be transferred to normobiotic recipients. In addition, I shared some preliminary data suggesting that butyrate, a potent inhibitor of histone deacetylases (HDACi), is a good candidate to influence the epigenome of HSPCs.
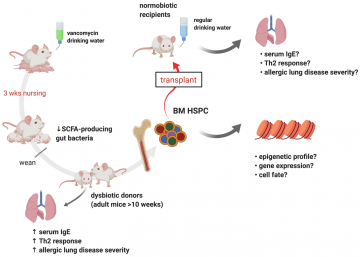
Figure 1. Experiments exploring the mechanisms linking the gut microbiome and allergic lung inflammation. Illustration made with Biorender.com
An epigenetic link between the gut microbiome and severity of allergic lung inflammation in mice
When thinking about how early life exposure to a specific microbiome and metabolites can influence long-term immunity, we considered which cell populations in mice (and humans) are responsible for long-term (or even lifelong) blood homeostasis. One obvious choice was long-term repopulating hematopoietic stem cells (HSCs). Our first experiment was to determine if we could transfer an “atopic high” phenotype from dysbiotic mice reared on vancomycin water to normobiotic mice reared on regular water by transplanting hematopoietic stem and progenitor cells (HSPCs). Using total bone marrow (BM) for this experiment, we found that we were indeed able to transfer an “atopic high” phenotype from dysbiotic mice to normobiotic recipients. Since we waited at least 10 weeks to check for signs of atopy in transplant recipients, this experiment suggests that BM progenitor cells from dysbiotic mice are altered by the microbiome (and their metabolites) with long-lasting consequences. Next, we needed to determine which BM progenitor cells convey these transplantable phenotypes and the molecular mechanisms that are responsible. At ASI 2019, I presented the above experiments and preliminary data suggesting that butyrate from SCFA-producing gut bacteria alters the epigenome of repopulating HSPCs. As a potent inhibitor of histone deacetylases (HDACi), butyrate is a good candidate to influence the epigenome of HSPCs. We will explore this hypothesis in subsequent experiments.
G’day MAIT!
There were two major themes at the ASI 2019 meeting (admittedly selected by my bias). There was a general theme of how the microbiome, particularly the gut microbiome, influences the pathology of several diseases in mouse models and human patients. For the most part, there was continued interest in the influence of gut microflora and metabolites on the usual suspects of mature effectors of immunity, including T cells, B cells, dendritic cells, and innate lymphoid cells. It is certainly critical for my work to keep up-to-date on how the gut microbiome influences the development, trafficking, and function of these immune cell lineages. The other theme of the meeting was a growing interest in mucosal-associated invariant T (MAIT) cells, which are non-conventional T cells that are restricted to recognize a major histocompatibility complex (MHC) class I-like surface protein (MR1) that presents vitamin B metabolites. For a recent MAIT cell review, see the publication by Godfrey et al (Nat Immunol 2019)4. MAIT cells help clear infection by pathogenic bacteria, fungi, and yeast that produce riboflavin and related metabolites. However, commensal microflora also likely influence MAIT cell expansion and survival, and they are implicated in the pathogenesis of allergic and autoimmune disease. Keeping in mind that there are important differences in the development and functional roles of these cells in mice and humans, MAIT cells have obvious implications for our work. It was inspiring to hear about this work from leaders in the field directly!
Acknowledgements
I would like to thank the CBR for the travel award that allowed me to present my work abroad. This project was initiated through a collaboration with Dr. William Mohn’s and Dr. Martin Hirst’s labs. Much of the background work was conducted by Dr. Alissa Cait (now a PDF at the Malaghan Institute of Medical Research in New Zealand). Misha Bilenky and Michelle Moksa of Dr. Hirst’s lab have provided expert contributions for epigenetic analysis for this project. I would also like to acknowledge Diana Canals Hernaez, Jessica Cait, and Kristen Li for their contribution to the work I presented at ASI 2019. Kristen Li and William Yip are continuing with this project in the Mohn and McNagny labs.
References
- Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152.
- Cait A, Cardenas E, Dimitriu PA, et al. Reduced genetic potential for butyrate fermentation in the gut microbiome of infants who develop allergic sensitization. J Allergy Clin Immunol. 2019;144(6):1638-1647 e1633.
- Cait A, Hughes MR, Antignano F, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018;11(3):785-795.
- Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20(9):1110-1128.


